IMD
Simulation of Vacuum Deposition of Atoms
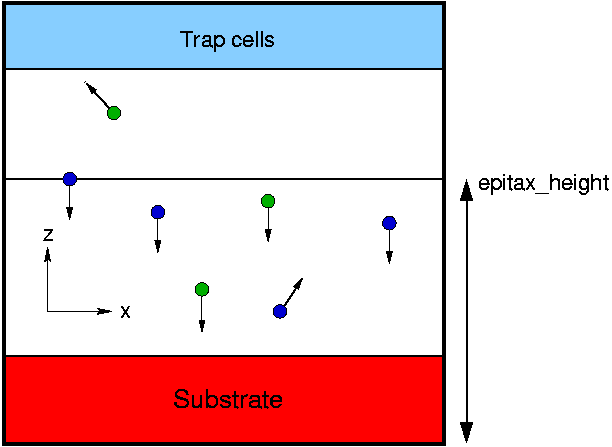
The option epitax enables the simulation of vacuum deposition of atoms on a substrate. During the simulation, a particle beam is created in the negative z direction (negative y direction in 2d) as shown in the figure. A substrate can be given as an atom input file. The default boundary conditions are periodic boundary conditions in the x and y direction (x direction in 2d) and free boundary conditions in z direction (y direction in 2d).
The creation of beam atoms begins at epitax_startstep (Default 0) and ends at epitax_maxsteps. The data for the atoms to be created must be given in the following form. The IMD parameter epitax_type contains the types of the atoms in a row. The masses of the atoms correspond to the parameter epitax_mass and have to be specified in a row corresponding to the atom types. The rate of creation (in time steps) of the atoms must be given in a row with the IMD parameter epitax_rate. The velocity of the atoms can be prescribed for each type as the corresponding temperature with the IMD parameter epitax_temp.
At epitax_startstep the atoms are created at the height given by the IMD paramter epitax_height. The x and y coordinate (x coordinate in 2d) are chosen randomly. However, the position is chosen in such a way that there is a distance larger than epitax_cutoff between the beam atoms. During the simulation, the height of creation of particles is moved upwards with a velocity corresponding to the IMD parameter epitax_speed.
Atoms in the beam are always integrated in the NVE ensemble, that is, their energy is conserved. The atoms in the substrate are in the ensemble specified by the IMD parameter ensemble as usual. Atoms that are deposited on the substrate are integrated with the substrate ensemble where the change of ensemble is performed as follows. At the beginning of the simulation, the mean potential energy Epot_mean of the atoms in the substrate is computed. The potential energy of the beam atoms initially being zero, the change of ensemble occurs if the potential energy of the beam atoms falls below C * Epot_mean where the control parameter C can be prescribed by the IMD parameter epitax_ctrl (Default 1).
Beam atoms that bounce off the substrate or atoms ablated from the substrate are removed from the simulation cell when they reach the top array of cells in the IMD cell decomposition (trap cells in the figure).
During the simulation, the atom number and the coordinates of the created atoms as well as the atom number and the z coordinates of the deleted atoms are given in the standard output.